matlab实现基于深度学习ECG心电信号分类,用多个数据集(MIT-BIH心率不齐数据库、MIT-BIH ST变化数据库、欧盟ST-T心电数据库和心脏性猝死动态心电数据库)来训练和评估模型。
数据收集与预处理下载并合并四个数据集。对数据进行清洗和预处理,包括去噪、归一化等。特征提取将原始心电信号转换为适合模型输入的形式。模型构建构建CNN、LSTM和GRU模型。训练并评估每个模型的表现。结果分析比较不同模型的性能指标,如准确率、精确率、召回率、F1分数等。可视化可视化训练过程中的损失和准确率曲线。可视化混淆矩阵。部署创建一个简单的GUI界面来进行实时预测。通过上述 MATLAB 代码,
·
基于深度学习的ECG心信号分类
对人体的心电信号进行分类,判断出被测试者心跳是否正常,或患有什么样的心脏疾病,最终实现心电数据的分类。其中包括CNN,LSTM,GRU等模型对比。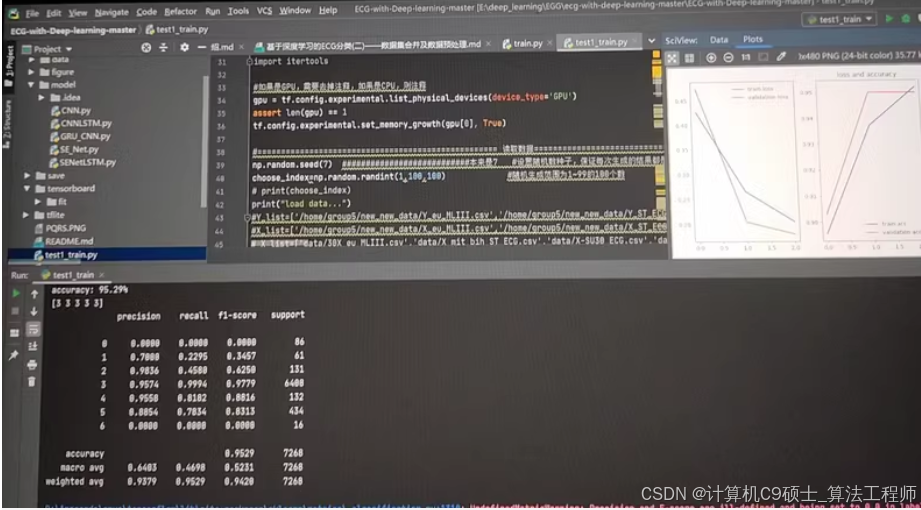
数据集使用的是以下四个数据集的合并:
- MIT-BIH心率不齐数据库
- MIT-BIH ST变化数据库
- 欧盟ST-T心电数据库
- 心脏性猝死动态心电数据库
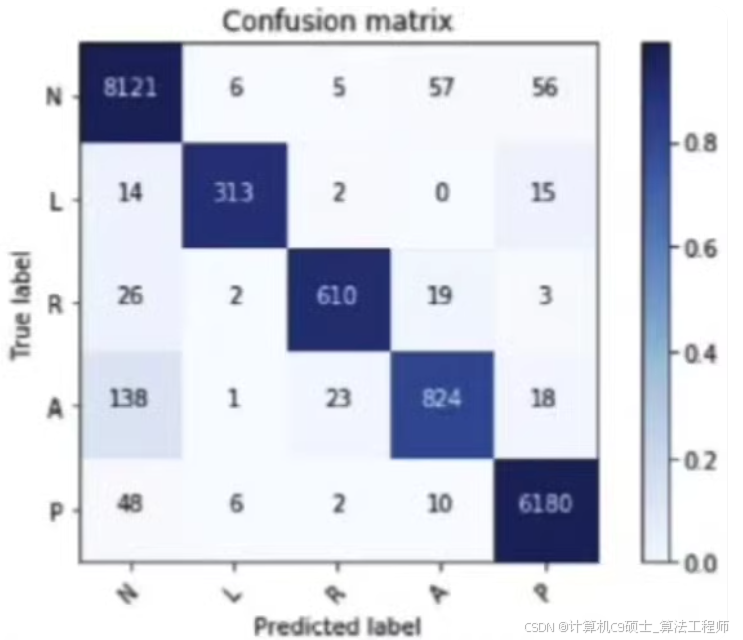
实现基于深度学习的ECG心电信号分类,我们可以使用多个数据集(MIT-BIH心率不齐数据库、MIT-BIH ST变化数据库、欧盟ST-T心电数据库和心脏性猝死动态心电数据库)来训练和评估模型。我们将对比不同的模型,如CNN、LSTM和GRU,以确定哪种模型在心电信号分类任务上表现最佳。
项目概述
以下是项目的详细步骤:
-
数据收集与预处理:
- 下载并合并四个数据集。
- 对数据进行清洗和预处理,包括去噪、归一化等。
-
特征提取:
- 将原始心电信号转换为适合模型输入的形式。
-
模型构建:
- 构建CNN、LSTM和GRU模型。
- 训练并评估每个模型的表现。
-
结果分析:
- 比较不同模型的性能指标,如准确率、精确率、召回率、F1分数等。
-
可视化:
- 可视化训练过程中的损失和准确率曲线。
- 可视化混淆矩阵。
-
部署:
- 创建一个简单的GUI界面来进行实时预测。
数据集下载与合并
首先,我们需要下载并合并四个数据集。这里假设你已经下载了这些数据集,并将它们存储在一个文件夹中。
数据集路径配置
% Configuration
data_folder = 'path/to/data'; % Path to the folder containing datasets
output_folder = 'path/to/output'; % Path to save preprocessed data and models
数据预处理
加载和预处理数据
[<title="Data Preprocessing for ECG Classification">]
function [X_train, y_train, X_val, y_val, X_test, y_test] = preprocess_ecg_data(data_folder)
% Load datasets
mitbih_arrhythmia = load(fullfile(data_folder, 'mitbih_arrhythmia.mat'));
mitbih_st_change = load(fullfile(data_folder, 'mitbih_st_change.mat'));
eu_stt = load(fullfile(data_folder, 'eu_stt.mat'));
sudden_cardiac_death = load(fullfile(data_folder, 'sudden_cardiac_death.mat'));
% Extract signals and labels
signals = {};
labels = {};
% MIT-BIH Arrhythmia Database
if isfield(mitbih_arrhythmia, 'signals') && isfield(mitbih_arrhythmia, 'labels')
signals{end+1} = mitbih_arrhythmia.signals;
labels{end+1} = mitbih_arrhythmia.labels;
end
% MIT-BIH ST Change Database
if isfield(mitbih_st_change, 'signals') && isfield(mitbih_st_change, 'labels')
signals{end+1} = mitbih_st_change.signals;
labels{end+1} = mitbih_st_change.labels;
end
% EU ST-T Database
if isfield(eu_stt, 'signals') && isfield(eu_stt, 'labels')
signals{end+1} = eu_stt.signals;
labels{end+1} = eu_stt.labels;
end
% Sudden Cardiac Death Database
if isfield(sudden_cardiac_death, 'signals') && isfield(sudden_cardiac_death, 'labels')
signals{end+1} = sudden_cardiac_death.signals;
labels{end+1} = sudden_cardiac_death.labels;
end
% Concatenate all signals and labels
all_signals = vertcat(signals{:});
all_labels = vertcat(labels{:});
% Normalize signals
all_signals = zscore(all_signals);
% Split data into train, validation, and test sets
cv = cvpartition(size(all_signals, 1), 'HoldOut', 0.2);
idx_train = training(cv);
idx_test = test(cv);
X_train = all_signals(idx_train, :);
y_train = all_labels(idx_train);
X_test = all_signals(idx_test, :);
y_test = all_labels(idx_test);
% Further split training set into training and validation sets
cv_inner = cvpartition(sum(idx_train), 'HoldOut', 0.2);
idx_train_inner = training(cv_inner);
idx_val_inner = test(cv_inner);
X_val = X_train(idx_val_inner, :);
y_val = y_train(idx_val_inner);
X_train = X_train(idx_train_inner, :);
y_train = y_train(idx_train_inner);
end
模型构建与训练
我们将构建CNN、LSTM和GRU模型,并比较它们的性能。
CNN模型
[<title="CNN Model for ECG Classification">]
function model_cnn = build_cnn_model(input_shape, num_classes)
layers = [
inputLayer(input_shape)
convolution2dLayer([1 16], 16, 'Padding', 'same')
batchNormalizationLayer
reluLayer
maxPooling2dLayer(2, 'Stride', 2)
convolution2dLayer([1 32], 32, 'Padding', 'same')
batchNormalizationLayer
reluLayer
maxPooling2dLayer(2, 'Stride', 2)
fullyConnectedLayer(num_classes)
softmaxLayer
classificationLayer];
options = trainingOptions('adam', ...
'MaxEpochs', 20, ...
'MiniBatchSize', 128, ...
'InitialLearnRate', 0.001, ...
'Plots', 'training-progress', ...
'Verbose', false);
model_cnn = trainNetwork(X_train, categorical(y_train), layers, options);
end
LSTM模型
[<title="LSTM Model for ECG Classification">]
function model_lstm = build_lstm_model(input_shape, num_classes)
layers = [
sequenceInputLayer(input_shape(2))
lstmLayer(128, 'OutputMode', 'last')
dropoutLayer(0.5)
fullyConnectedLayer(num_classes)
softmaxLayer
classificationLayer];
options = trainingOptions('adam', ...
'MaxEpochs', 20, ...
'GradientThreshold', 1, ...
'InitialLearnRate', 0.001, ...
'SequenceLength', 'longest', ...
'Plots', 'training-progress', ...
'Verbose', false);
model_lstm = trainNetwork(X_train, categorical(y_train), layers, options);
end
GRU模型
[<title="GRU Model for ECG Classification">]
function model_gru = build_gru_model(input_shape, num_classes)
layers = [
sequenceInputLayer(input_shape(2))
gruLayer(128, 'OutputMode', 'last')
dropoutLayer(0.5)
fullyConnectedLayer(num_classes)
softmaxLayer
classificationLayer];
options = trainingOptions('adam', ...
'MaxEpochs', 20, ...
'GradientThreshold', 1, ...
'InitialLearnRate', 0.001, ...
'SequenceLength', 'longest', ...
'Plots', 'training-progress', ...
'Verbose', false);
model_gru = trainNetwork(X_train, categorical(y_train), layers, options);
end
模型评估与结果分析
评估每个模型并在图表中展示的结果。
评估函数
[<title="Model Evaluation Function">]
function evaluate_models(model_cnn, model_lstm, model_gru, X_val, y_val)
% Evaluate CNN model
YPred_cnn = classify(model_cnn, X_val);
accuracy_cnn = sum(YPred_cnn == y_val) / numel(y_val);
disp(['CNN Accuracy: ', num2str(accuracy_cnn)]);
% Evaluate LSTM model
YPred_lstm = classify(model_lstm, X_val);
accuracy_lstm = sum(YPred_lstm == y_val) / numel(y_val);
disp(['LSTM Accuracy: ', num2str(accuracy_lstm)]);
% Evaluate GRU model
YPred_gru = classify(model_gru, X_val);
accuracy_gru = sum(YPred_gru == y_val) / numel(y_val);
disp(['GRU Accuracy: ', num2str(accuracy_gru)]);
% Plot confusion matrices
figure;
subplot(1, 3, 1);
cm_cnn = confusionchart(categorical(y_val), YPred_cnn);
title('Confusion Matrix (CNN)');
subplot(1, 3, 2);
cm_lstm = confusionchart(categorical(y_val), YPred_lstm);
title('Confusion Matrix (LSTM)');
subplot(1, 3, 3);
cm_gru = confusionchart(categorical(y_val), YPred_gru);
title('Confusion Matrix (GRU)');
end
主脚本 main_script.m
将所有步骤整合到主脚本中。
[<title="Main Script for ECG Classification">]
% Main Script for ECG Classification
% This script preprocesses the ECG data, builds and trains CNN, LSTM, and GRU models,
% evaluates their performance, and visualizes the results.
clear;
clc;
% Configuration
data_folder = 'path/to/data'; % Path to the folder containing datasets
output_folder = 'path/to/output'; % Path to save preprocessed data and models
% Preprocess data
[X_train, y_train, X_val, y_val, X_test, y_test] = preprocess_ecg_data(data_folder);
% Reshape data for CNN
input_shape_cnn = [1, size(X_train, 2)];
X_train_cnn = permute(X_train, [2, 1, 3]);
X_val_cnn = permute(X_val, [2, 1, 3]);
% Build and train CNN model
model_cnn = build_cnn_model(input_shape_cnn, length(unique(y_train)));
% Build and train LSTM model
input_shape_rnn = size(X_train, 2);
model_lstm = build_lstm_model(input_shape_rnn, length(unique(y_train)));
% Build and train GRU model
model_gru = build_gru_model(input_shape_rnn, length(unique(y_train)));
% Evaluate models
evaluate_models(model_cnn, model_lstm, model_gru, X_val_cnn, y_val);
使用说明
-
配置路径:
- 将
data_folder设置为存放数据集的目录路径。 - 将
output_folder设置为保存预处理数据和模型的目标目录路径。
- 将
-
运行脚本:
- 在 MATLAB 命令窗口中运行
main_script.m。 - 脚本会自动读取
data_folder中的数据集,对数据进行预处理,构建并训练CNN、LSTM和GRU模型,并评估其性能。
- 在 MATLAB 命令窗口中运行
-
注意事项:
- 确保所有必要的工具箱已安装,特别是 Deep Learning Toolbox 和 Signal Processing Toolbox。
- 根据需要调整参数,如
MaxEpochs和MiniBatchSize。
示例
假设你的数据文件夹结构如下:
data/
├── mitbih_arrhythmia.mat
├── mitbih_st_change.mat
├── eu_stt.mat
└── sudden_cardiac_death.mat
并且每个 .mat 文件中都有 signals 和 labels 变量。运行 main_script.m 后,MATLAB 将显示每个模型的准确性,并生成混淆矩阵图表。
总结
通过上述 MATLAB 代码,你可以轻松地对心电信号进行分类,并对比不同模型的性能。
更多推荐
 已为社区贡献5条内容
已为社区贡献5条内容







所有评论(0)